News!
eyeIntegration version 1.0 went live early this year (2019-01-16) and recently was accepted for publication in IOVS. In celebration of the news, I’m posting a small series of posts about the genesis, development, upgrades, and future of eyeIntegration.
You can find our latest manuscript on bioRxiv. The latest update should go live soon.
Background
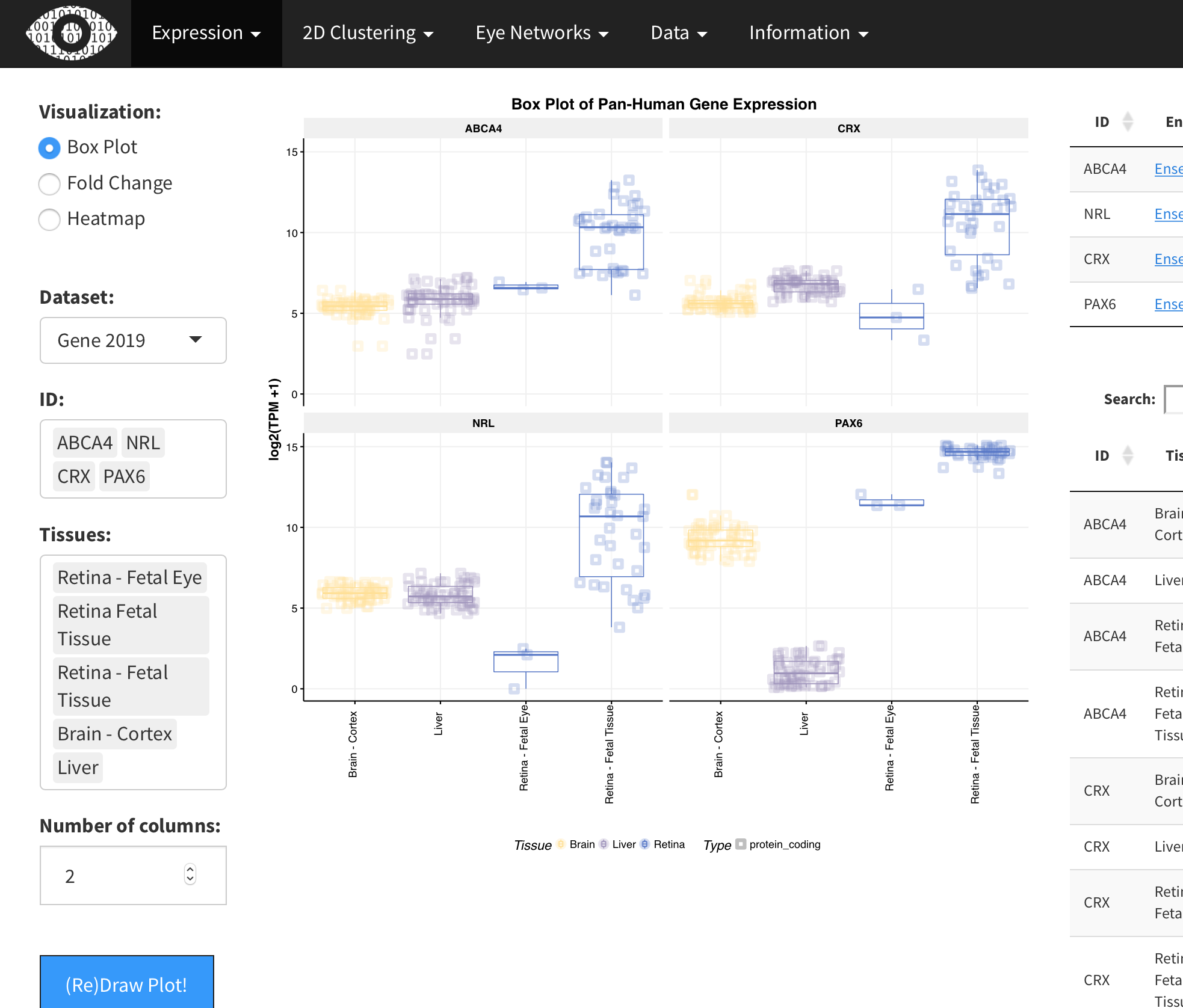
eyeIntegration was developed to serve as a quick and easy to use normal gene expression portal in eye tissues.
 The inspiration came from GTEx, which contains dozens of
post-mortem tissue types from hundreds of samples (and samples are people who have donated their
body to science - thanks!).
The inspiration came from GTEx, which contains dozens of
post-mortem tissue types from hundreds of samples (and samples are people who have donated their
body to science - thanks!).
Knowing what the normal gene expression is a crucial piece of information when doing novel gene function research. You are going to be a lot less excited about an interesting gene if you also know it has zero expression in the retina.
Since no eye tissues are collected by GTEx, I had to build my own version. The best solution would involve setting up a very expensive study - with a carefully designed tissue collection, DNA/RNA extraction, etc. protocols along with huge amounts of paperwork required when working with people. This would also be very expensive and take multiple years.
As I have approximately zero research dollars, I did the next best thing and just grabbed all the publicly available RNA-seq eye samples from GEO / SRA / ENA.
Since I knew R and did not know any web development stuffs I chose to use Shiny to build the web portal. In 2016 (my first commit was May 24th, 2016), when this project got started this seemed sort of nuts to do this. But the documentation was pretty good and it wasn’t too hard to spin up a Shiny server on a linux box I had under my desk. Over the past three years RStudio has continued to develop Shiny and I’m feeling better and better about going this route.
What do you mean by One Developer Portal?
I work for the National Eye Institute NEI, as a staff scientist (not a PI/professor - which means I don’t have a pot of $ to do research). I provide bioinformatic support / data analysis for a branch in NEI.
eyeIntegration is a essentially a side-project I’ve done in my “spare time” and is run by myself (with important features written by two post-bacs , John Bryan and Vinay Swamy). It is possible to make and support a resource like this because of available compute and great software.
The long-term goal is to keep eyeIntegration current (e.g. take in new samples) with part-time effort from one person (me) with periodic bursts of activity to add new features.
This is getting sort of long so I’m going to break it into several pieces:
- Making a performant site
- What’s new with eyeIntegration 1.0? (future post)
- Crucial software bits (future post)
- What will eyeIntegration 2.0 be? (future post)