This post is a continuation from here.
Really important stuff I learned to make a performant web site in Shiny
After a few months of tinkering I had a working web app on my local computer, which
is a 32GB of RAM, 1TB SSD Mac Pro trashcan. All of the data objects were
.Rdata, which were load() when the site was initialized. This was fine
in the beginning and in fact the shiny site was deployed with this structure
in May of 2017. I fairly quickly realized that this was a really bad idea.
As there are 20,000+ genes and I was storing the gene expression information for hundreds of samples, this was a sort of large data set (about 20e6 total data points).
On my computer I could get the site started up in about 15 seconds or so. On the slower server it took upwards of 45 seconds to initialize the site. Which was a problem as the Shiny Server puts a site to “sleep” whenever there’s a lack of activity of around 5 minutes (this can be changed in your own Shiny Server). Plus it was using a LOT of memory and when I tried to load the site at home, on my not insanely fast internet connection, it would take a minute or so to get the site running.
I then discovered the crucial tool Profvis which allows you to benchmark exactly how much time each function / process is taking on a Shiny site. Here I learned two important (and in hindsight, super obvious) things:
- Loading hundreds of megaabytes
.Rdataobjects takes a while (this was 90% of the slow site initializing time) - Having the
ui.R(browser) side load gene lists (vectors which are 20,000+ in length) and big images just kills people with slow internet connections
Solving Problem 1: .Rdata objects
Using .Rdata to store the objects was clearly a bad idea as it made the site
slow to load and made it impractical to further expand the data set (e.g. more samples and
transcript-level information).
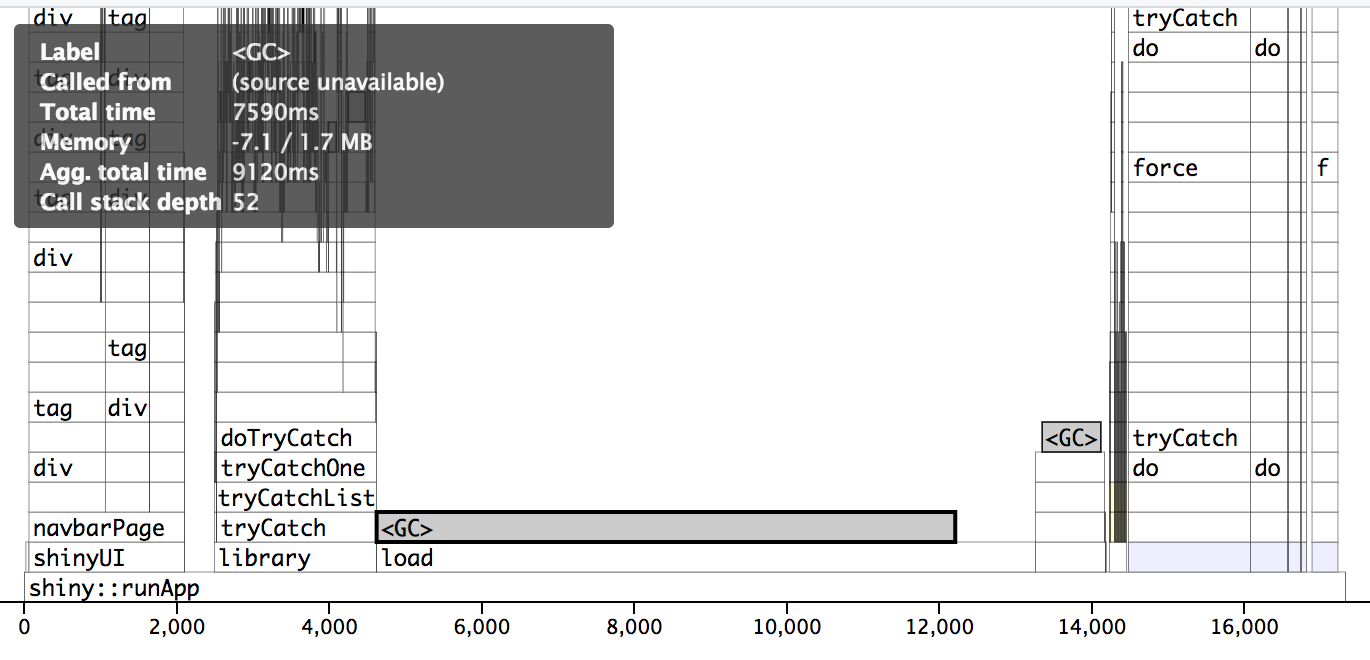
Profvis pre-optimization. The wall/actual time is 38 seconds. Notice how long the load section is on a fast SSD. This was substantially worse on the actual server.
So I need some sort of data structure that could store the information in a way that R could access it without holding the whole thing in memory. The answer I chose was SQLite. Why not some other database structure? Well, my needs were fairly simple - one web site that reads one database. I’d also dabbled with SQLite before and found it pretty simple to write SQL queries.
It ended up being pretty simple to implement, as RStudio has put substantial effort in getting SQLite (and other database types) to interface with R.
The biggest problem was that I originally tried to just replicate my data structure, which was organized with samples (>1000) as columns and genes (>20,000) as rows
SQLITE_MAX_COLUMN was set to 1000, I think. So a straight copy didn’t work. After
some Googling I realized I was being an idiot and SQLite databases
were supposed to be LONG.
So I restructured the database to have three columns:
- Gene name
- Sample name
- Gene expression value
This isn’t disk space efficient (especially since SQLite can’t be compressed!) but that’s not a concern as disk is cheap.
After restructuring the server.R file to use the SQLite, the initializing time
dropped from ~38 seconds to ~5 seconds!
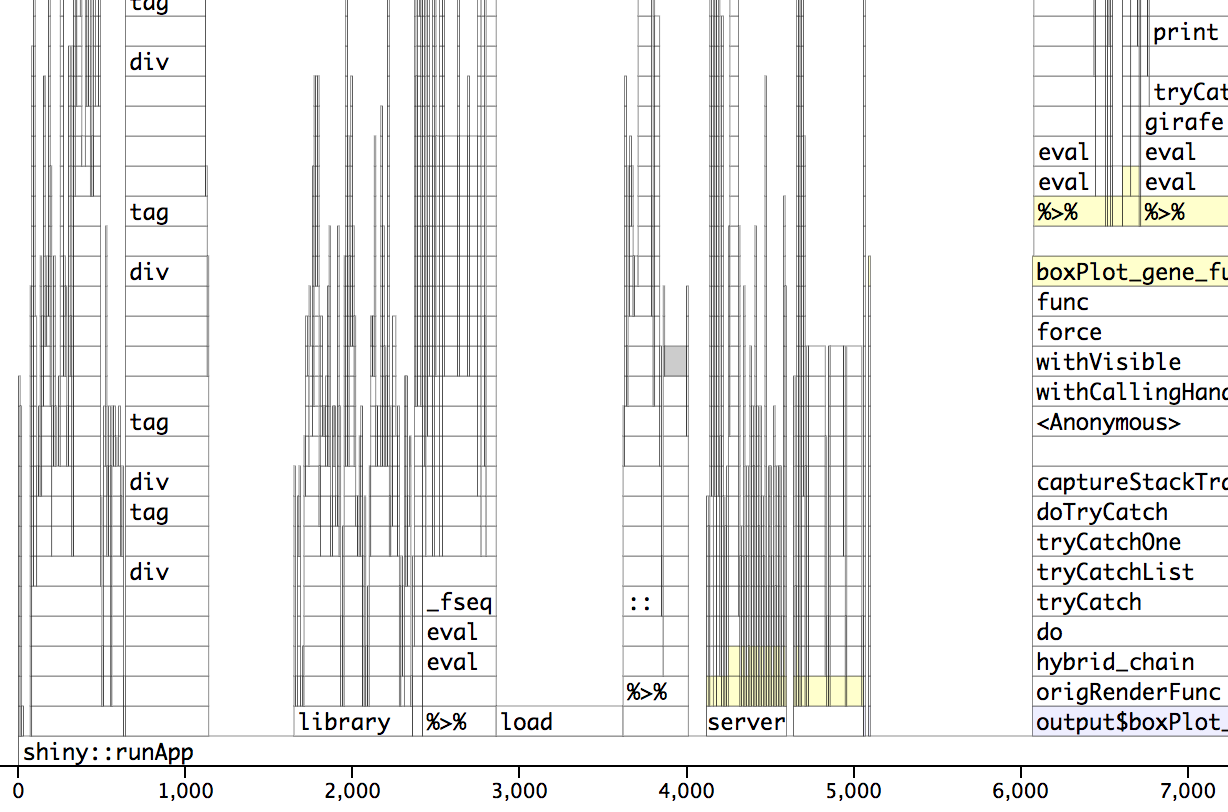
Profvis post-optimization. The wall/actual time is 5 seconds. See how the load section is now about 1 second.
Solving Problem 2: Slow load times with slow internet
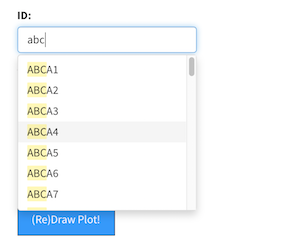 The
The updateSelectizeInput function provides user selectable from a drop-down list.
I use this to allow the user to pick a gene of interest to get expression information.
Problem is that a 20,000 long vector is a few megabytes - which has to be re-downloaded
to the user’s browser EVERY TIME THEY VIST THE SITE.
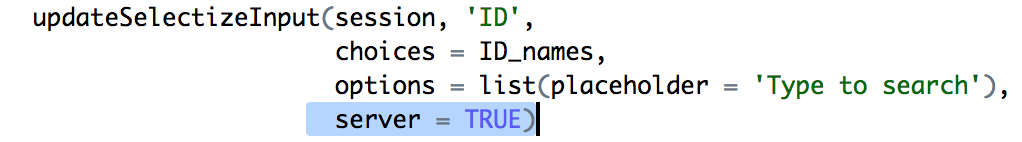
The crucial bit.
Fotunately the Shiny authors also realized that this could be a problem and provided
an option (server) in updateSelectizeInput which shifts the burden of holding
the list to the server. This was implemented pretty quickly by having the server
load the gene list and setting server = TRUE.
After doing this and removing some stupid large images the site load time went from tens of seconds (!) to instaneous with fast internet and a couple of seconds with slower internet.